Utilising Semantic Web Ontologies to publish Experimental Workflows
Workshop on Enabling Decentralised Scholarly Communication (SciEDSC) - co-located with Extended Semantic Web Conference (ESWC)
✍ Harshvardhan J. Pandit* , Ensar Hadziselimovic , Dave Lewis
Description: Reviewing workflow reproducibility using semantic web and their publication with traditional documents along with licensing annotations
published version 🔓open-access archives: harshp.com , TARA , web (canonical) , zenodo
📦resources: code , Utilising Semantic Web Ontologies to publish Experimental Workflows
Abstract
Reproducibility in experiments is necessary to verify claims and to reuse prior work in experiments that advance research. However, the traditional model of publication validates research claims through peer-review without taking reproducibility into account. Workflows encapsulate experiment descriptions and components and are suitable for representing reproducibility. Additionally, they can be published alongside traditional patterns as a form of documentation for the experiment which can be combined with linked open data. For reproducibility utilising published datasets, it is necessary to declare the conditions or restrictions for permissible reuse. In this paper, we take a look at the state of workflow reproducibility through a browser based tool and a corresponding study to identify how workflows might be combined with traditional forms of documentation and publication. We also discuss the licensing aspects for data in workflows and how it can be annotated using linked open data ontologies.
Keywords
Table of Contents
Introduction
A key tenet of scientific research is the sharing of experiment data. Publications offer a decentralised way to gauge validity of research by utilising the collective wisdom of the community through peer review. The increasing demand for open access means researchers must share details about their experiment such as implementation steps and datasets in a highly accessible and structured manner. Traditional patterns of publication such as journals are reacting to this demand by providing increasingly interactive access to data that is often embedded or displayed along with the published paper. However, such methods of publication do not take into consideration the reproducibility of the experiment as an important metric which puts the onus of ensuring sufficient resource sharing and access on the researchers who largely fail to take it into consideration.
Reproducibility in scientific experiments allows other researchers to reproduce the experiment to obtain results that can confirm or dispute the original claims [1]. To encourage verifiability and adoption of methods, access to the original experiment and results along with its components or datasets must be provided in a transparent and declarative manner. Research published through the peer-review process is seen as having credibility for its correctness which does not reflect upon its reproducibility. Approaches such as attributing source code via online repositories such as Github or executable components through Docker or Virtual Machines help share the technology behind the experiment, though this creates additional problems due to the sheer diversity in differing technologies and frameworks in the software world.
Workflows capture complex methods and their interactions as a series of steps [2] and have been used successfully in several different areas of scientific research [3,4,5]. There have been several efforts to map workflows as linked data ontologies [a,b,c,d] along with several tools and frameworks that help users in publishing workflows. As workflows encapsulate the experiment and its subsequent execution, they are also useful in assessing the reproducibility of research by including them in publications.
Workflows can be helpful in defining and sharing experiments along with associated resources using linked open data principles which can help streamline the process and make them more accessible. We aim to investigate means to discern the parity between adoption of workflows as a documentation mechanism and determining how researchers carry out research documentation and the associated challenges in augmenting existing publication mechanisms using linked open data principles. To this aim, we have modelled an experiment to better understand documentation habits and publication challenges for workflows and data licenses using a browser based tool. We also present a discussion of the current state of affairs and the need for a more decentralised model of publication that augments traditional approaches.
The rest of the paper is laid out as follows: In Section 2, we discuss the background and related work with respect to workflows and data licensing. We explain the motivation for identifying workflow documentation through a browser based tool in Section 3, with the licensing aspect of datasets discussed in Section 4. We conclude our discussion in Section 5 with an outlook towards future work.
Background & Related Work
Capturing Provenance in Experiment Workflows
Provenance is information about entities, activities, and people (or software) involved in producing data or a component which can be used to form an assessment about its quality, reliability, or trustworthiness. The PROV ontology, which is a W3C recommendation since 30th April 2013, provides definitions for interchange of provenance information. Using PROV, we can define entities and the various relations and operations between them such as generated by, derived from, and attributions. PROV has been successfully utilised in several domains and applications [e] including encapsulation of scientific workflows [6,7] and provenance repositories [8,9].
PROV was designed to be generic and domain independent, and needs to be extended to address the requirements to represent workflow templates and executions [10]. P-Plan extends PROV to represent plans that guide execution of scientific processes and describes how the plans are composed and their correspondence to provenance records that describe the execution itself. OPMW re-uses the Open Provenance Model core vocabulary along with extending both PROV and P-Plan to describe workflow traces and templates. OPMW is mostly suited as an ontology to describe workflows in a manner aligning with how researchers design and conduct experiments, and has been used in tools and frameworks to capture experimental workflows.
OPMW allows representation of workflows at a very granular level. In OPMW, a workflow template represents the design of the workflow containing different steps or processes. Artifacts are part of a template and are used or generated by the processes. There are two types of artifacts - data variables and parameter variables. Data variables can be used as inputs and can also be generated by processes whereas parameters work as expected for workflow steps. OPMW reuses terms from Dublin Core to represent attribution for author, contributor, rights and license of datasets and the code used in the workflow. Workflow Executions are bound to the template and represent an execution run. Each step or process in the template has a corresponding execution process linked to it containing provenance statements about its execution. Execution Artifacts used or generated during execution are linked to their corresponding artifact from the template. Executions have terms used to define the start and end of execution traces along with metadata for artifacts such as file location, file size, and declaration of agents that perform or are involved in the execution process such as scripts, or tools used to design and/or execute workflows.
There are several tools that allow the creation and consumption of workflows [11,12,13,14]. WINGS [15] is an end-to-end workflow system that allows describing and instantiating high-level workflow templates and executing them in various executing environments. It uses an implementation of OPMW to model workflows into templates and executions and stores them as a catalogue and features workflow reuse. Workflows can utilise data variables from the catalogue while parameters are limited to literal values. WINGS can interleave metadata generated during execution to utilise it in workflow design and processes which allows creation of partial workflows that can be incrementally iterated towards completion and execution.
A related tool called WorkflowExplorer allows navigating workflow templates along with their metadata and execution results. It displays information as a webpage consisting of all resources related to the template grouped by their common type and retrieves this data dynamically. Each resource is a link to a webpage describing it and shows information about it such as if an execution run has been successful or listing execution instances for a template variable. Another tool for documentation of workflows is the Organic Data Science Wiki, which can generate persistent documentation for workflows automatically from the repository.
Workflow fragments can be described as a collection of workflow components which form a subset of the workflow and represent some distinct functionality. Fragments can be shared at a more granular level than workflows, and can thus be reused more easily. Experiments that utilise the same fragments can be linked or clustered based on their metadata, though such experiments would not necessarily be constituted as variations of a common template. The idea of enacting reproducibility over such fragments rather than the workflow as a whole has seen some interest [16].
Reproducibility
Reproducibility is the ability to reproduce the results of an experiment with the goal to confirm or dispute the experiments claims [1]. It requires access to the description of the original experiment and its results along with workflows that capture the different settings required to accurately reproduce the execution environment. The terms repeatability and variation are commonly aligned with reproducibility whose formal definitions can be found in [17]. Reproduction of experiments is based on availability of resources which may not be accessible or were changed since the experiment execution. Reproducibility in such cases becomes challenging as comparing workflows between the original and a rerun is non-trivial and time-consuming.
Research Objects [18,19] encompass initiatives that allow the bundling together of all resources and metadata associated with an experiment. Each resource is identified using a globally unique identifier such as DOI for publication or ORCID for researchers. Resource objects can aggregate information related to workflows such as original hypothesis, inputs used in executions, and workflow definitions along with execution traces of workflow runs. Annotations attached to the research object can include provenance traces and information about workflow evolution and its component elements. TIMBUS Context Model [20] is similar in aims as Research Objects while additionally allowing bundling of legal metadata such as copyright licenses and patents and intellectual property rights. Its authors have presented a mapping from Context Model to Research Object making them compatible in usage and consumption. VisTrail [14] allows creation of reproducible papers that contain description of the experiment, links for input data, applications, and visualisations for the execution outputs. ReproZip can help with capturing provenance information along with any environmental parameters required for execution into a self-contained reproducible package.
Previously mentioned approaches that mitigate these problems look at capturing all the information required to define and reproduce an experimental workflow. As this information often contains datasets, resources, and services which can change or become inaccessible, the associated workflows can no longer be successfully shared or utilised. In [21], the authors evaluate workflows and term this phenomenon as ‘workflow decay’. They analysed 92 Taverna workflows and list four causes of workflow decay which are missing volatile third party resources, missing example data, missing execution environment, and insufficient description about workflows. In [22], the authors examined 613 papers from ACM conferences, out of which 515 contained tools developed by the authors themselves, 231 contained accessible source code of which only 123 could be successfully built. Common causes of failure were missing environment variables and incorrect or unspecified dependencies. In another comprehensive study [23], the authors analysed nearly 1500 workflows from the myExperiment repository that used Taverna. They found that 737 workflows were accessible and executable workflows, out of which 341 executed without errors while only 29.2% of 1443 datasets were usable.
Reproducibility challenges and best practices has seen several discussions. In [23,24], the authors present six strategies for creation of reproducible scientific workflows that focus on defining and sharing of all information and data in a clear and persistent manner. [25] discusses the best practices for workflow authors with a particular focus on how to prevent workflow decay. The various challenges in workflow reproducibility arising from third party services is discussed in [26,27]. In [25] the authors present seven types of (meta-)data required to make workflows reproducible of which some needs to be defined manually by the user, while the rest can be inferred from provenance data or generated automatically by the system. In [28] the authors define two types of reproduction - physical and logical. Physical reproducibility conserves workflows by packaging all its components so that an identical replica can be created and reused, whereas logical reproducibility requires workflows and components to be described with enough information for others to reproduce a similar workflow in future. [29] uses this principles to utilise Docker as a workflow environment that packages the experiment execution and services along with required data.
In [29,30], the authors investigate the probability of making a workflow reproducible. They use decay parameter [31] which is the probabilistic term used to define four categories of reproduction based on their probability for reproducibility, which are reproducible, reproducible with extra cost, approximately reproducible, reproducible with probability P, and non-reproducible. The authors also present operational definitions for various terms based on the decay parameter. Repeatability is executing the experiment again (in exactly the same manner) with the same environmental and user specific parameters where the decay parameters are any randomly values such as system noise or captured timestamps. Variability is where the workflow is run on the same infrastructure with some intentional modification of the jobs. Portability is repetition in a different environment and reproducibility is defined as being a combination of repeatable and portable.
By considering provenance traces as acyclic graphs, it is possible to utilise graph analysis to find relationships and interactions between workflows. Data artifacts or activities are considered as nodes with the links denoting relationships between them. By tracing data flow in a graph, it is possible to reflect and infer the production and consumption of data for workflow executions. PDIFF [1] utilises this approach to determine whether an experiment has been reproduced by identifying points of divergence between graphs of differing workflows. It tries to find if the two workflows represent the same execution trace, and if they do not, then at what point do they diverge. FragFlow [32] is another approach utilising graphs to obtain workflow fragments that relate workflows to each other and indicate parts that are more likely to be reused. In [33], the authors present a technique to reduce visual complexity in workflow graphs. They argue that the visualisation generated by combining the logical and structural attributes leads to a better understanding of complex and relatively unfamiliar systems.
Along with approaches that focus on enabling the creation and consumption of research, there has been a growing discussion on the principles and methods used in the publication and reproduction of workflows along with associated resources such as datasets. The Joint Declaration of Data Citation Principles [34] states that data should be machine readable and treated the same as papers in a scholarly ecosystem. The FAIR Principles [35], which stand for findable, accessible, interoperable and reusable data, encourage semantic interoperability through reuse of data. Linked Research [36] defines the requirements for a web-based ecosystem for scholarly communication which makes it possible to publish links to workflows and other related resources using existing technologies. LERU Roadmap for Research Data [37] recommends identifying documentation and metadata requirements at the start of a project which would then comply with existing standards for the content. It also advocates creation, processing and sharing of data with the scientific community through a generic framework for a wide variety of research processes and outputs. OpenAIRE aims to substantially improve the discoverability and reusability of research publications and data by interconnecting large-scale collections of research outputs across Europe. The central idea for the project is to create workflows and services on top of repository content to form an interoperable network which can act as an all-purpose repository which would be open for all researchers.
Reproducibility Enhancement Principles (REP) [38] is a set of recommendations based on the Transparency and Openness Promotion (TOP) guidelines along with other discussions regarding data publication amongst funding agencies, publishers, journal editors, industry participants and researchers. REP argues that access to the computational steps taken to process data and generate findings is as important as access to data themselves which lends to the argument about publishing workflows and its associated resources. The authors consider the ability to reproduce an experiment through its steps on the same data as the original authors as a minimum dissemination standard. This includes the workflow information describing the resources and its relationship to the steps used in computation of the results. It also suggests that journals should conduct a reproducibility check as part of the publication process and should enact the TOP standards at level 2 or level 3 which would ensure that all data and code is available persistently in an open trusted repository.
There has been discussion [40] into weaker forms of reproducibility where rather than replicating an entire workflow, only a few parts or components of it are fashioned to be reusable. While workflow fragments are ideal for such scenarios, it still undermines the difficulties that may arise in its reproduction due to a variety of reasons such as technical configuration or data availability and licensing. Additionally, traditional mechanisms of publication do not address these challenges in any meaningful way, which restricts the possibility of a centralised solution. Recent advances into decentralising this process [36] allows publication of research in an open and accessible format without funnelling it into centralised research repositories. Tools that help consume and annotate published papers can also be extended to reflect workflows and components for the same experiment. As the decentralisation process allows the researcher to hold sufficient control over the layout and contents of the published research, it can be utilised as a gateway in the interest of reproducibility.
We extend our argument based on these recommendations to discuss various means of disseminating existing knowledge amongst researchers to try and identify possible drawbacks in existing approaches and to discover ways in which traditional approaches in conducting research can benefit from LOD principles and workflow based systems.
Licensing
When it comes to publishing the datasets, there are many different variables that need to be considered. First is the need for context regarding limitations on publication such as public or intra-institution [41]. This should be complemented with the mode of access describing where the data is stored and availability regarding how it can be accessed. There needs to be a clear strategy about licensing and whether it applies to a subset or the complete data. This is vital in cases where data can potentially contain personal or sensitive information. There are established mechanisms and providers for data publishing in academic circles such as Mendeley Data, PLOS, and Dryad.
It is necessary to have a deeper understanding of the licensing issues along with laws and policies that may be applicable. This includes defining rules pertaining to the intellectual property (IP) of the assets and relevant privacy policies. Without clear understanding of what is freely available to be reproduced in an observed dataset, it is very difficult to know which data is permissible to be accessed and under which conditions can it be used. There needs to be an effective mechanism to check the status of intellectual property or licensing issues that might arise in the process. This includes integrity of the research ethics undertaken in conducting the original experiment that produced the data along with replication and generating more datasets.
Due to the nature of linked open data, it is possible to see how information related to experimental workflows can be effectively interlinked without a centralised mechanism. What remains is to find and utilise appropriate models for declaration of legalities associated with data. Best practices for publishing linked data, authored by W3C, states that licenses should be explicitly connected to the data itself. This allows for a transparent definition of the circumstances under which a third-party can reuse the datasets. Creative Commons (CC) is the suggested approach for licensing associated with such declarations.
There are two main mechanisms to describe and communicate the permissions of a dataset. The first is a license which is a legal instrument for rights holder to permit certain operations over data to other parties [42]. The second mechanism is a waiver which in practice is enforced as giving up the ability to claim rights over to other parties. Commonly used conditions in licensing models are attribution, copyleft, and non-commerciality. Attribution is giving the original author credit for the work on operations such as distributing, replicating, and displaying. Copyleft assumes that the derived work must use the same licensing model as the work it is derived from. Non-commercial clauses stipulate usage for non-commercial applications except under specified conditions.
Datasets are subject to so-called attribution stacking, meaning all of the contributors to the original work must be attributed in the chain of production. As a derived work may include datasets under different licensing models, all of the derivatives authors and licences must be taken into consideration when producing the final licensing model.
Licensing of datasets is a very complex issue when it comes to publishing experimental data. Most of the licensing mechanisms including CC are primarily designed to protect the published work and not necessarily the datasets. There are ongoing efforts to address this issue. Open Data Commons (ODC) is a set of legal tools that help provide and use Open Data with ODC Open Database License (ODbL) that relates to publishing of datasets. Science Commons, which is now merged with Creative Common under the Open Science initiative that specifically targets the use of data in scientific environment.
There are currently only a few options available to evaluate data from a legal perspective. While there are certain mechanisms that assess licensing and IP issues, specifically META-SHARE licenses, the actual usage is limited based on the context of the data and need for a manual assessment.
The idea is to have all the assets in the experiment tied to certain licenses and possibly graded to describe their level of openness for repeatability and reusability. This is achievable using a Rights Expression Language (REL) which is an ontology to express rights using linked data. Open Digital Rights Language (ODRL) is a REL developed to express rights, rules, and conditions including permissions, prohibitions, obligations, and assertions and the rules pertaining to IP issues. ODRL can be used to expand existing ontologies to contextualise experimental data through the use of its own semantic vocabulary. However, there needs to be an awareness of any potential limitations of using the ODRL language to determine complexities in licensing issues.
ODRL has an expressive vocabulary that makes it possible to explain
permission-related relationships in a precise manner. Examples are
‘grantUse’, ‘annotate’, ‘reproduce’
permissions and many more.
Additionally, ODRL has the concept of permission inheritance that enables
granting of permissions to dependent variables based on permissions inherited
from independent variables (arguments) of the experiment. It has both XML and
JSON based schema for easier integration and implementation.
There can be multiple assets, assigners, and assignees associated with permission models that describe permissions, prohibitions, duties, and constraints. All the attributes can be inherited as well as passed on to another party. Translating all of this to an experimental workflow use case, it is possible to deal with an experiment’s licensing models and permission inheritance for only certain fragments or the entire experiment. Through this a privacy policy can be clearly set that defines a retention policy along with any IP details that can be passed using parent-child relationships to executions or variations of that experiment.
Browser based tool for workflow documentation
We created a browser based tool as a test-bed for our discussion and study of the current methods for workflow documentation and publication. The focus of the tool was in advancing knowledge about the use of vocabularies in facilitating sharing and repeatability of experiments and replication of results. The tool also focused on the workflow documentation and its role in publication of the experiment and subsequent discovery of related work. We focused on researchers in areas aligned with Natural Language Programming (NLP) and Machine Learning (ML) as these contain a good variety of variations in experiment workflows where executions are highly interlinked and repetitive by nature. Additionally, there have been a number of previous approaches and ontologies [43] targeting these specific areas which provides motivation for further discussion. The target audience for the study is researchers not primarily familiar with linked open data vocabularies for describing experimental workflows.
Prospective participants are first asked to fill in a questionnaire (termed pre-questionnaire) to gauge their familiarity with experimental workflows and linked open data. The pre-questionnaire enquires about experience in sharing workflows and whether the participants are familiar with the concepts of reproducibility and workflow reuse. Academic qualification along with published research is used as a metric of experience and familiarity with the research area. The questionnaire also seeks to understand experiences of researchers in using a variation of existing or prior work. This is enquired through questions about the use of a slight or small modification of previous research, either from self or other researchers. The pre-questionnaire can be found online at here.
We chose OPMW as the target vocabulary for describing workflows as it allows experimental workflows to be described in a highly descriptive manner by capturing steps, datasets and their relationships. Rather than asking users to learn the ontology, or in some cases, the concept and use of linked open data, we abstract use of the specification and focus on the documentation aspect of workflows. Users of the tool are not required to know the underlying use of OPMW to use the tool, but are presented with simplified concepts and structure from the ontology. The explicit use of terms and metadata used to define and describe resources which can be searched or explored is provided as the basis of the system. They are provided with the general idea of a template being an abstract design of the experiment which contains steps and datasets interlinked to define control flow. These templates can then be instantiated into multiple executions each containing distinct outputs and resources similar to the notion of a generic experiment run. Users are also exposed to how workflows can be documented using the information provided and linked with related resources.
The documentation generated within the tool follows the principles of linked open data where each resource has its own corresponding properties and attributes. For e.g. an execution instance will contain links to every resource it is associated with, such as the template it was based on, its execution processes and artifacts along with their corresponding template parameters, steps, and data variables. This allows a comprehensive overview of the entire workflow as well as the ability to follow these links to the documentation for a particular resource.
The tool, which can be accessed here, is hosted on an internal virtual machine hosted by Trinity College Dublin running in a python virtual environment. For the server side, it uses flask as the web framework and rdflib for interacting with RDF data. As rdflib is backend-agnostic, and to keep the tool footprint small for an online demonstration, we use an SQLite single-file serverless database as a triple-store. On the client side, it uses standard web technologies along with some additional libraries and JointJS for rendering the workflow as a graph. It contains a few useful features for testing and the study such as importing and exporting workflows using JSON which allows workflows to be loaded or saved from within the tool. This is particularly useful for the study as it allows users to interact with partially filled workflows by simply importing the corresponding JSON.
The experiment contains three tasks, which combined together can take about one hour in terms of time for completion. To test the tool and the underlying study, we propose that users be assigned one task based on their familiarity with workflow documentation and running executions. This can be gauged by analysing their response to the pre-questionnaire. Users who are not familiar with linked open data or with using workflows can start with Task 1 which asks them to search for experiments containing specified attributes and resources using a form based interface. For users who are familiar with experimentation practices and workflows, Task 2 requires completion of an execution for an existing template. Task 3 can be suited for users who are familiar with linked open data and publication of workflows or are experienced with the concepts of reproduction and repeatability. The task asks them to create a variation of an existing template as an example of modifying existing research. Each tasks targets a different aspect of workflow documentation and consumption. Although the three tasks are disjoint with each other, they all converge on the documentation generated for the workflows which the users are encouraged to explore at the end of their task.
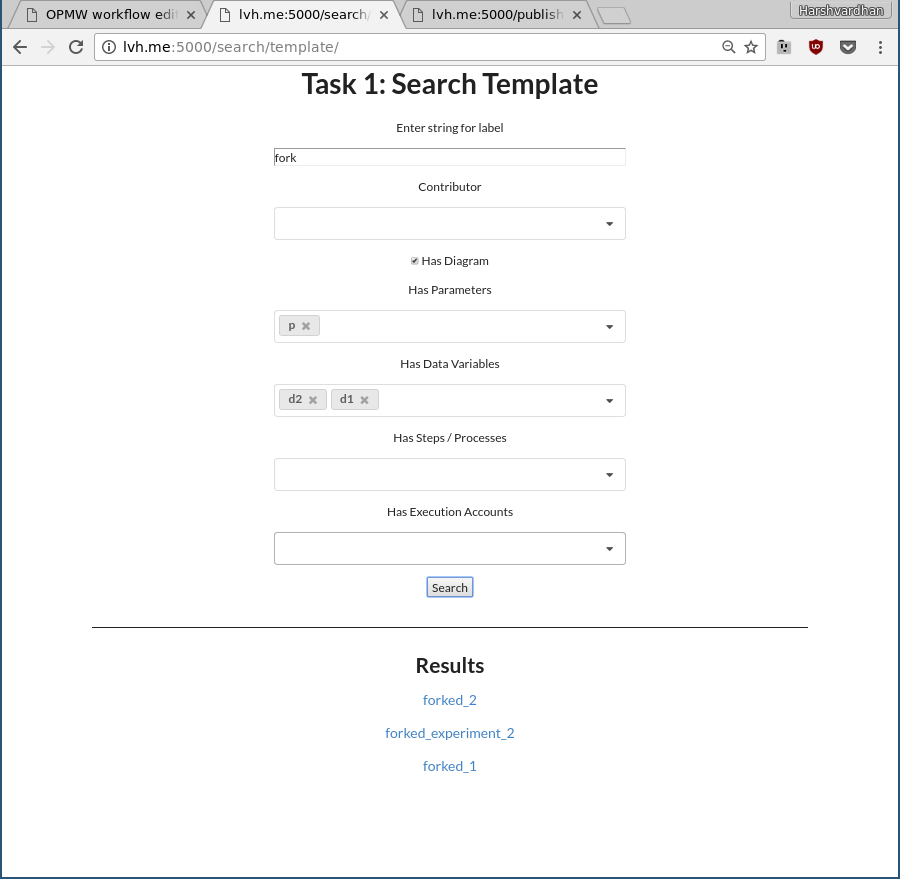
In Task 1, the user is asked to search for experiments containing the specified attributes and resources. The form based interface (see Fig. 1) allows specifying the search parameters using a combination of fields for each attribute and resource such as specifying a substring in the template name, having certain author(s), containing a particular step or dataset, or based on template executions. Based on the arguments supplied, the tool returns workflow templates that contain or match the given criteria which are shown at the bottom in the form of hyperlinks. The user is asked to explore the results produced by the query to know more about a particular experiment and its execution runs and variations. This task asks the user to think about workflows as being documented using metadata for itself as well as all of its resources and the advantages of being able to filter or link together queries based on this information. It also exposes them to workflow documentation and the way different experiments and resources can be linked or explored in an automatically generated documentation. Internally, the tool uses a SPARQL query to retrieve templates.
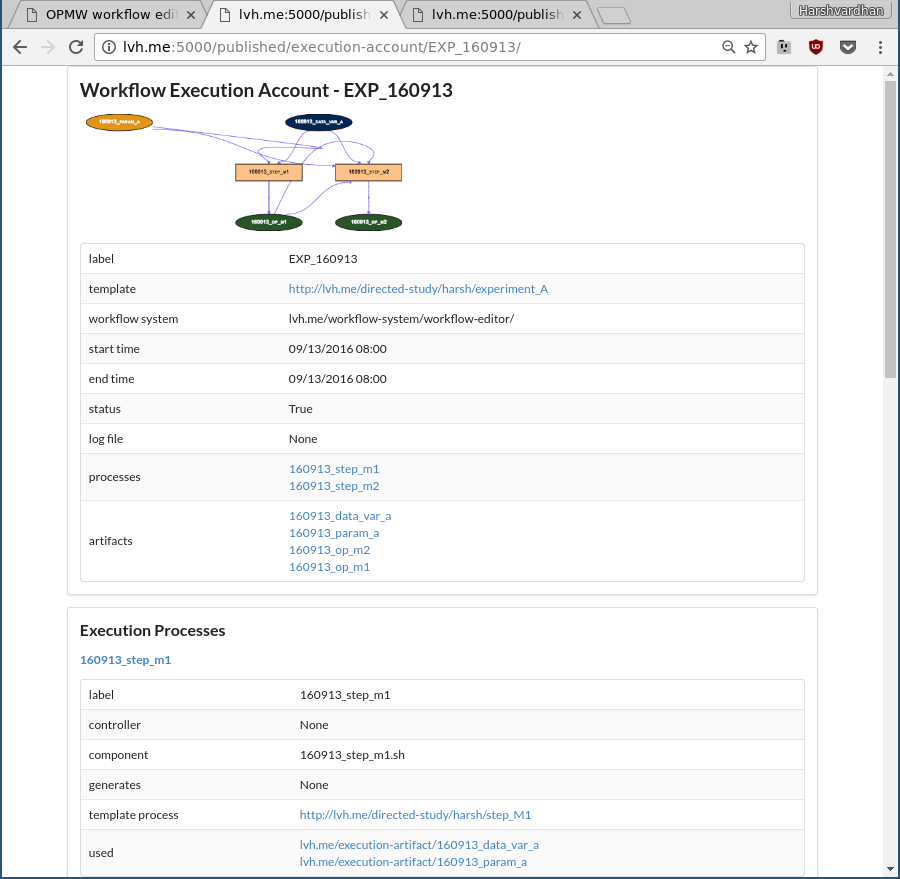
Task 2 involves the user completing a partially complete execution for an existing template. Users need to fill in the missing metadata which for steps could be the author information if it was a researcher or a software agent for scripts along with recording the step's starting and finishing time. For datasets the missing metadata can be the location URI or whether the dataset is stored as a file or a folder. The tool shows appropriate errors or warnings until the required information is correctly filled after which the workflow is published and saved in the triple-store. Users are then asked to view documentation (see Fig. 2) generated for their execution. Following displayed links allows users to explore things such as other executions for the same template, executions run by the same author or agent or utilising the same datasets. The task allows users to interact with a workflow system that can follow execution runs and collect them under a common experiment template. Users also see an example of how a dataset can be linked to multiple executions through the use of an URI. The idea of storing experiment results in this manner and the subsequent collection of execution runs allows users to discover execution runs or experiments with the desired results. As there are no specific instructions given to the users regarding the working of the tool, any method of discovery or exploration is based on their understanding of how workflows are linked together. This is deliberate owing to the nature of linked open data and the open world assumption.
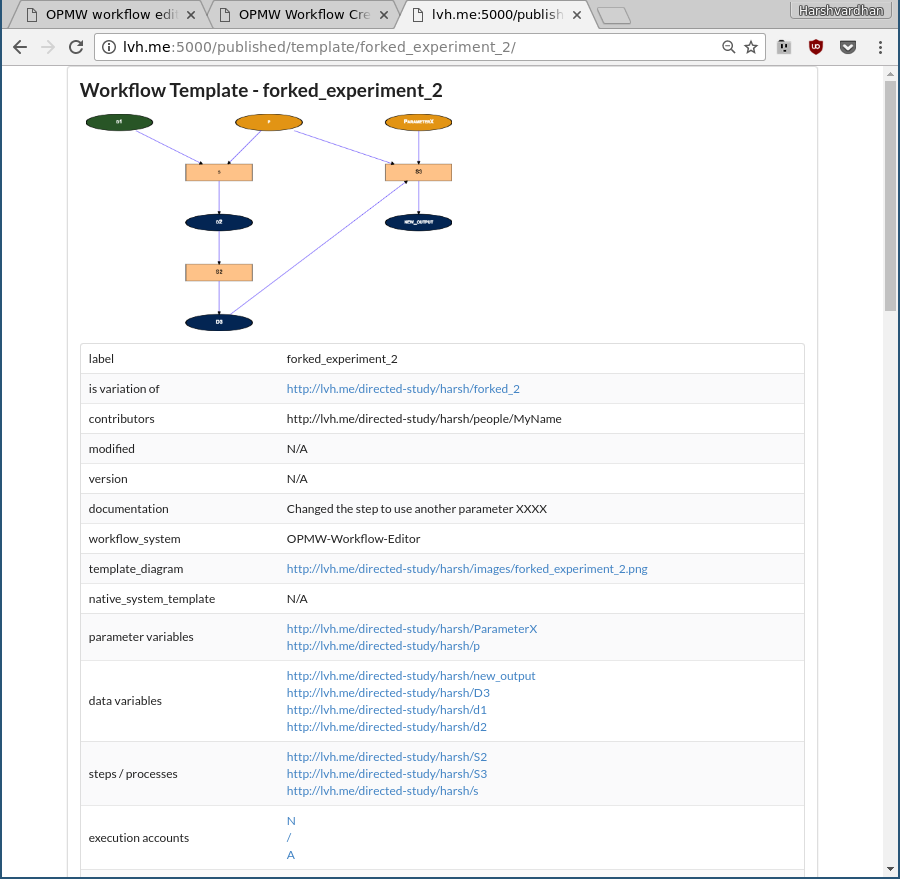
Task 3 asks the user to create a variation of an experiment by modifying an existing template. Examples provided for variation are modifying an existing step by changing the datasets and parameters it uses or adding new steps and/or datasets to modify the control flow. As the notion of variation is vague and ambiguous, users will not be given concrete instructions in terms of what constitutes a variation and are free to modify the experiment as long as it can still be sufficiently comparable to the original template. Upon successful completion, they are shown the documentation for the template along with a description and link to the original template which listed their variation of the experiment (see Fig. 3). The task helped users discover variations of experiments that could potentially show alternate approaches towards the same goal. The executions of each variation are only associated with that particular template and are not shared with the original. This allows a possible query by the user to see which variation produced the desired results and under which (parametric) conditions.
As OPMW does not specify any term we can use for denoting that a template
is a variation of another template, we introduced a placeholder term isVariationOf
based on prov:wasDerivedFrom
and prov:wasRevisionOf.
It associates two templates together as being variations
but does not specify which resources are shared or what exactly has been modified. Ideally,
any ontology specifying such variation should also be expressive enough to describe
what resources in the workflow have been changed or are affected by the change.
The example specified template labelled forked2 as the variation
of the template step123. More work needs to be done in this area to
specify degree of variance between experiements and to express the nuances between
variation, forking, and iteration of experiment templates.
@prefix this_project: <http://lvh.me/directed-study/workflow/> .this_project:forked_2 a opmw:WorkflowTemplate,prov:Plan ;rdfs:label "forked_2" ;this_project:isVariationOf this_project:steps123 ;
The use of OPMW and the styling of documentation is inspired from previous research and workflow tools such as WINGS and WorkflowExplorer that show a description of the experiment along with all of its properties and resources which can be navigated using the hyperlinks. For templates and executions, the tools shows a graphical representation of the steps and artifacts as a visualisation to help the user understand the structure of the experiment. The steps and artifacts are structured as nodes on the graph with connections between them depicting control flow. Each type of resource is depicted in a visually distinct manner so that it is easy to differentiate them. The documentation is generated by interpreting the underlying RDF graph as a webpage with resources linked using hyperlinks. Where possible, additional information is displayed about resources to encourage discovery of related items. For example, a step described in an experiment template contains entries for all templates it is used in which can be clicked to access the documentation page for that template.
After the end of the given task(s), a post usage questionnaire (post-questionnaire) is used to evaluate the responses of the participants. It contains open ended questions about the usefulness of the tool for workflow documentation and in exploration and discovery of existing research components. The post-questionnaire also enquired about their views on incorporating such workflow tools in their existing research work. At the end of the session, a non-structured and optional interview is conducted to help better understand the responses for qualitative responses such as how they plan to incorporate linked open data or workflows into their existing research. The post-questionnaire along with the optional interview is useful to form views about challenges faced in incorporating workflows as means of documentation and publication and whether there are any significant areas of concern in its adoption. These discussions are also helpful in understanding the state of affairs in publication of experiment data and how it can be combined with the linked open data principles. A link to the online post-questionnaire can be found here.
Licensing workflow resources
Rights expression languages can be used to describe the serialisation of data relating to an IP or privacy policy. One of the main challenges in this process are licensing issues related to data, methods and assets used in the experiments. Depending on how these resources are licensed, the repeatability of the experiment changes along with the conditions for reuse. If not declared properly, utilising published research data can become burdened with legal issues. Therefore, it is crucially important to evaluate the current state regarding researchers’ understanding of licensing related to publications and experimental workflows.
There are two contexts that must be observed in understanding the licensing process: one is regarding the entire workflow as a whole, and the other is specific parts of the workflow, including but not limited to some of the steps, algorithms, or datasets. Producing an appropriate license for experimental workflows thus poses a challenge as licenses are not necessarily ‘sum of parts’, but each part has to be considered in its own contexts. Additionally, the workflow has to be analysed in a more precise manner regarding licenses that apply in a local or regional legislation or have patent and ownership issues.
It is possible to produce a grading depicting the potential for reusability for resources. This can be done by focusing on individual parts of the workflow and placing them into the above mentioned contexts, summarising gathered relationships and inheritance, and then producing the final grading model. Datasets in experimental workflows can be annotated using a schematic based on colour such as red depicting unavailability for reuse. For the purpose of simplicity, we will be discussing annotating only the experimental datasets although annotations can be applied to any experimental resource falling under the licensing policy.
Expanding on ODRL, there are two distinct approaches for annotating workflows depending on whether the experiment is in the process of being initiated (original experiment, original data), or reproduced. In the first case, authors of the experiment are looking to publish the original work and need to find an appropriate license under which to do so. In latter case, the person repeating or reusing the experiment would like to understand what the attached licenses mean in terms of publication for a derivative work. Annotation can thus go both ways, whether explaining the attached licenses’ implications, or suggesting a new licensing model.
We discuss here the use of ODRL as the ontology used for describing licenses associated with workflows. First step would be determining the context for the experimental data being annotated. Authors responsible for creating the original experiment and dataset are termed as Assigner, with the Assignee denoting the person(s) repeating the experiment. Keywords used match ODRL concepts, with the most important ones being use, attribute, and reproduce. As there are limited options for licensing datasets, most licenses can be covered by using CC and ODC licenses. Assignees analyse the attached license and express required conditions using ODRL. Analysis of the aggregated concepts then produces the grading which identifies warnings or alerts based on usage. Terms and keywords like pay, sell, obtain consent would raise a warning flag whereas watermark, translate, shareAlike would raise an alert flag. Other cases and conditions can have different flags depending on their own contexts.
By using ODRL, it is possible to have a system that identifies potential legal issues surrounding data availability for sharing and using data. Annotations make licenses and issues easier to apprehend by non-legal parties through a visual grading of resources. Utilising a colour based grading allows the flexibility to differentiate based on flow or usage of data and whether the person(s) in question are the original authors or replicators. There are still some challenges in applying the ontology to specific instances of an experimental workflow. Some terms used by ODRL are ambiguous in their meaning or similar to other terms whereas some terms might not be applicable at all. Through a subset or a possible extension the ontology can tackle the vast majority of use cases in the real life scenarios of experimental practice.
Conclusion & Future Work
Adopting linked open data for dissipating experiment workflows opens new opportunities for dissemination of knowledge. Sharing workflows helps reproducibility of experiments as a core issue with publication along with access to experiment data and resources. By combining these with documentation efforts for experiment authors, we discussed how research can be better disseminated and shared towards the advancement of science.
We adopted OPMW as ontology for describing workflows along with ODRL for declaring licensing to create a workflow tool based in the browser. The tool acts as the central theme for discussions with researchers and allows them to interact with experiments via the generated documentation and to explore existing research. By abstracting away the underlying ontology, users focus on consumption of workflows and the exploration or related research through documentation. The tool along with the associated discussions and questionnaires allows us to evaluate the state of workflow publishing for researchers not explicitly familiar with workflow ontologies and data licensing using linked open data principles. We are currently evaluating user studies and responses based on the tool with a focus on its documentation aspects.
Our main aim in terms of forming this study and the development of the tool was in understanding the overlap between current workflow and documentation habits, particularly for NLP and ML researchers. By studying current documentation habits and available linked open data ontologies, we hypothesised a tool through which users can be exposed to workflows created with OPMW. The study associated with the tool looks towards indentifying areas where linked open data adoption can be simplified and incorporated into traditional forms of publications. We also discuss licensing using ODRL for annotating the experiment workflow and datasets. Licensing workflows and datasets is important for reproducible workflows, as it lays out the conditions under which the experiment may produce further work or be evaluated. We tried to envision a novel approach for integrating licensing in workflow documentaion. One idea we found potentially useful was color-coding based on suitability for reuse, and would like to emphasise this idea for potential future work.
In terms of future work, we would like to further enhance the tool using various state of the art research approaches that can help in furthering our discussions into workflow documentation. We particularly would like to emphasise the use of graph analysis to differentiate between experiments to identify and highlight variations and help the user visually interact with them. As OPMW does not currently have terms associated with variation, there is an opportunity for an extension to be created addressing the interlinking of related workflows. We would also like to investigate the means of publishing workflows in a decentralised manner using linked data. The possibility of enabling researchers to host their workflows themselves while providing a central repository information about executing it could potentially be helpful in increasing reproducibility analysis for published workflows. Such information could then be attached with the published papers as annotations that can guide the users to updated information on the workflows rather than letting them decay. Another thing we would like to evaluate is making it easier for researchers to provide documentation in a way close to how they conduct experiments, and to bundle this together in a publication.
The tool tries to visualise experiment workflows for users and generates documentation based on OPMW to describe workflows and resources. However, some users prefer working with other forms of documentation that do not align well with linked open data or formal forms of publication. An example can be keeping notes in markup languages such as markdown where there is a distinct structure to the document but no formal keywords to add context. It may be possible to look into utilising such text based styles to document experiments by converging them using ontologies such as OPMW. This would allow users the choice of using tools or writing their own documentation which can then be converted into linked open data.
Along with access to papers and experimental workflows, the data associated with the experiment must also be made available in the interest of reproducibility and furthering research. Such datasets should have licenses that declare the terms under which the data was obtained and the conditions under which it may be accessed or re-used. A common example in research publications is the condition where experimental data may only be re-used in an academic environment, expressly forbidding any commercial usage. Such clarity in license is beneficial and essential for research as it allows access to a large corpus of shared data that can help in future experiments as well as in reproducibility of previous research. In cases where such data cannot be made available, publication of its schema can allow researchers to utilise the experiment or its components by compiling a matching dataset. The schema in such a case would correspond to metadata pertaining to the dataset that describes what kind of data it encapsulates and how it is structured without exposing any of the actual data itself. Such approaches are helpful in experiments where personalised data is often anonymised and may not be released under any permissible license. We would like to explore this issue through the use of a grading mechanism utilising ODRL within the workflow tool.
Acknowledgements
This work has been supported by the European Commission as part of the ADAPT Centre for Digital Content Technology which is funded under the SFI Research Centres Programme (Grant 13/RC/2106) and is co-funded under the European Regional Development Fund.
References
- P. Missier, S. Woodman, H. Hiden, and P. Watson, ‘Provenance and data differencing for workflow reproducibility analysis: PROVENANCE AND DATA DIFFERENCING FOR REPRODUCIBILITY’, Concurrency and Computation: Practice and Experience, vol. 28, no. 4, pp. 995–1015, Mar. 2016.
- Y. Gil, ‘Intelligent workflow systems and provenance-aware software’, in Proceedings of the Seventh International Congress on Environmental Modeling and Software, San Diego, CA, 2014.
- J. Ruiz, J. Garrido, J. Santander-Vela, S. Sánchez-Expósito, L. Verdes-Montenegro, ''AstroTaverna: Building workflows with Virtual Observatory services', Astron. Comput., 7–8 (2014), pp. 3–11 Special Issue on The Virtual Observatory: I
- I.D. Dinov, J.D.V. Horn, K.M. Lozev, R. Magsipoc, P. Petrosyan, Z. Liu, A. MacKenzie-Graham, P. Eggert, D.S. Parker, A.W. Toga 'Efficient, distributed and interactive neuroimaging data analysis using the LONI Pipeline', Frontiers in Neuroinformatics, Volume 3 (2009)
- K. Wolstencroft, R. Haines, D. Fellows, A. Williams, D. Withers, S. Owen, S. Soiland-Reyes, I. Dunlop, A. Nenadic, P. Fisher, J. Bhagat, K. Belhajjame, F. Bacall, A. Hardisty, A.N. de la Hidalga, M.P.B. Vargas, S. Su, C. Goble 'The Taverna workflow suite: designing and executing workflows of web services on the desktop, web or in the cloud', ' Nucleic Acids Res. (2013)
- K. Belhajjame, J. Zhao, D. Garijo, M. Gamble, K. Hettne, R. Palma, E. Mina, O. Corcho, J.M. Gómez-Pérez, S. Bechhofer, G. Klyne, C. Goble 'Using a suite of ontologies for preserving workflow-centric Research Objects', Web Semant. Sci. Serv. Agents World Wide Web (2015)
- P. Missier, S. Dey, K. Belhajjame, V. Cuevas-Vicenttín, B. Ludäscher 'D-PROV: Extending the PROV provenance model with workflow structure', Proceedings of the 5th USENIX Workshop on the Theory and Practice of Provenance, TaPP’13, USENIX Association, Berkeley, CA, USA (2013), pp. 9:1–9:7
- Víctor Cuevas-Vicenttín, Parisa Kianmajd, Bertram Ludäscher, Paolo Missier, Fernando Chirigati, Yaxing Wei, David Koop, Saumen Dey 'The PBase scientific workflow provenance repository', Int. J. Digit. Curation, 9 (2) (2014), pp. 28–38 View Record in Scopus | Citing articles (2)
- Khalid Belhajjame, Jun Zhao, Daniel Garijo, Aleix Garrido, Stian Soiland-Reyes, Pinar Alper, Oscar Corcho, 'A workflow PROV-corpus based on taverna and WINGS', in: Proceedings of the Joint EDBT/ICDT 2013 Workshops, Genova, Italy, 2013, pp. 331–332.
- D. Garijo, Y. Gil, and O. Corcho, ‘Abstract, link, publish, exploit: An end to end framework for workflow sharing’, Future Generation Computer Systems, Jan. 2017.
- I.D. Dinov, J.D.V. Horn, K.M. Lozev, R. Magsipoc, P. Petrosyan, Z. Liu, A. MacKenzie-Graham, P. Eggert, D.S. Parker, A.W. Toga 'Efficient, distributed and interactive neuroimaging data analysis using the LONI Pipeline', Frontiers in Neuroinformatics, Volume 3 (2009)
- J. Goecks, A. Nekrutenko, J. Taylor 'Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences', Genome Biol., 11 (8) (2010)
- K. Wolstencroft, R. Haines, D. Fellows, A. Williams, D. Withers, S. Owen, S. Soiland-Reyes, I. Dunlop, A. Nenadic, P. Fisher, J. Bhagat, K. Belhajjame, F. Bacall, A. Hardisty, A.N. de la Hidalga, M.P.B. Vargas, S. Su, C. Goble 'The Taverna workflow suite: designing and executing workflows of web services on the desktop, web or in the cloud', Nucleic Acids Res. (2013)
- F. Chirigati, J. Freire, D. Koop, C. Silva, 'VisTrails provenance traces for benchmarking', in: Proceedings of the Joint SDBT/ICDT 2013 Workshops, 2013, pp. 323–324.
- Y. Gil, V. Ratnakar, J. Kim, P.A. Gonzalez-Calero, P.T. Groth, J. Moody, E. Deelman 'WINGS: Intelligent workflow-based design of computational experiments', IEEE Intell. Syst., 26 (1) (2011), pp. 62–72
- Harmassi M., Grigori D., Belhajjame K. (2015) 'Mining Workflow Repositories for Improving Fragments Reuse'. In: Cardoso J., Guerra F., Houben GJ., Pinto A., Velegrakis Y. (eds) Semantic Keyword-based Search on Structured Data Sources. Lecture Notes in Computer Science, vol 9398. Springer, Cham
- Vitek, Jan, and Tomas Kalibera. "R3: Repeatability, reproducibility and rigor." ACM SIGPLAN Notices 47, no. 4a (2012): 30-36.
- Sean Bechhofer, John Ainsworth, Jitenkumar Bhagat, Iain Buchan, Phillip Couch, Don Cruickshank, Mark Delderfield, Ian Dunlop, Matthew Gamble, Carole Goble, Danius Michaelides, Paolo Missier, Stuart Owen, David Newman, David De Roure, Shoaib Sufi (2013) 'Why Linked Data is Not Enough for Scientists', Future Generation Computer Systems 29(2), February 2013, Pages 599-611, ISSN 0167-739X, doi:10.1016/j.future.2011.08.004
- Khalid Belhajjame, Jun Zhao, Daniel Garijo, Matthew Gamble, Kristina Hettne, Raul Palma, Eleni Mina, Oscar Corcho, José Manuel Gómez-Pérez, Sean Bechhofer, Graham Klyne, Carole Goble (2015) 'Using a suite of ontologies for preserving workflow-centric research objects', Web Semantics: Science, Services and Agents on the World Wide Web, doi:10.1016/j.websem.2015.01.003
- R. Mayer, T. Miksa, and A. Rauber, ‘Ontologies for Describing the Context of Scientific Experiment Processes’, 2014, pp. 153–160.
- J. Zhao et al., ‘Why workflows break—Understanding and combating decay in Taverna workflows’, in E-Science (e-Science), 2012 IEEE 8th International Conference on, 2012, pp. 1–9.
- Moraila, Gina, Akash Shankaran, Zuoming Shi, and Alex M. Warren. 'Measuring Reproducibility in Computer Systems Research'. Tech Report, 2014.
- R. Mayer and A. Rauber, ‘A Quantitative Study on the Re-executability of Publicly Shared Scientific Workflows’, 2015, pp. 312–321.
- Piccolo, Stephen R., and Michael B. Frampton. "Tools and techniques for computational reproducibility." GigaScience 5, no. 1 (2016): 30.
- A. Bánáti, P. Kacsuk, and M. Kozlovszky, ‘Minimal sufficient information about the scientific workflows to create reproducible experiment’, in Intelligent Engineering Systems (INES), 2015 IEEE 19th International Conference on, 2015, pp. 189–194.
- D. De Roure, K. Belhajjame, P. Missier, J. M. Gómez-Pérez, R. Palma, J. E. Ruiz, K. Hettne, M. Roos, G. Klyne, and C. Goble, “Towards the preservation of scientific workflows,” in Proceedings of the 8th International Conference on Preser- vation of Digital Objects (iPRES 2011), Singapore, 2011.
- K. Belhajjame, C. Goble, S. Soiland-Reyes, and D. De Roure, “Fostering scientific workflow preservation through discovery of substitute services,” in E-Science (e-Science), 2011 IEEE 7th International Conference on, Dec 2011, pp. 97–104.
- Santana-Perez, Idafen, Rafael Ferreira da Silva, Mats Rynge, Ewa Deelman, María S. Pérez-Hernández, and Oscar Corcho. "Reproducibility of execution environments in computational science using Semantics and Clouds." Future Generation Computer Systems 67 (2017): 354-367.
- A. Bánáti, P. Kárász, P. Kacsuk, and M. Kozlovszky, ‘Evaluating the average reproducibility cost of the scientific workflows’, in Intelligent Systems and Informatics (SISY), 2016 IEEE 14th International Symposium on, 2016, pp. 79–84.
- A. Bánáti, P. Kacsuk, and M. Kozlovszky, ‘Evaluating the reproducibility cost of the scientific workflows’, in Applied Computational Intelligence and Informatics (SACI), 2016 IEEE 11th International Symposium on, 2016, pp. 187–190.
- A. Banati, P. Kacsuk, M. Kozlovszky, M. 'Four level provenance support to achieve portable reproducibility of scientific workflows'. In Information and Communication Technology, Electronics and Microelectronics (MIPRO), 2015 38th International Convention on IEEE. unpublishe
- Garijo, Daniel, Oscar Corcho, Yolanda Gil, Boris A. Gutman, Ivo D. Dinov, Paul Thompson, and Arthur W. Toga. "Fragflow automated fragment detection in scientific workflows." In e-Science (e-Science), 2014 IEEE 10th International Conference on, vol. 1, pp. 281-289. IEEE, 2014.
- T. Koohi-Var and M. Zahedi, ‘Linear merging reduction: A workflow diagram simplification method’, in Information and Knowledge Technology (IKT), 2016 Eighth International Conference on, 2016, pp. 105–110.
- Callaghan, Sarah. "Joint declaration of data citation principles." (2014).
- Wilkinson, Mark D., Michel Dumontier, IJsbrand Jan Aalbersberg, Gabrielle Appleton, Myles Axton, Arie Baak, Niklas Blomberg et al. "The FAIR Guiding Principles for scientific data management and stewardship." Scientific data 3 (2016).
- https://linkedresearch.org/
- P. Ayris, R. D. W. Group, and others, ‘LERU Roadmap for Research Data’, 2013.
- V. Stodden et al., ‘Enhancing reproducibility for computational methods’, Science, vol. 354, no. 6317, pp. 1240–1241, 2016.
- Roure DD, Belhajjame K, Missier P, Al E. 'Towards the preservation of scientific workflows'. Proceedings of the 8th International Conference on Preservation of Digital Objects (iPRES 2011), Singapore, 2011; 228–231.
- Cohen-Boulakia S, Leser U. 'Search, adapt, and reuse: the future of scientific workflows'. SIGMOD Record 2011; 40(2):6–16. DOI: http://doi.acm.org/10.1145/2034863.2034865.
- Alexander, Keith, Richard Cyganiak, Michael Hausenblas, and Jun Zhao. "Describing Linked Datasets." In LDOW. 2009.
- Ball, A. (2014). ‘How to License Research Data’. DCC How-to Guides. Edinburgh: Digital Curation Centre. (2011)
- McCrae, John P., Penny Labropoulou, Jorge Gracia, Marta Villegas, Víctor Rodríguez-Doncel, and Philipp Cimiano. "One ontology to bind them all: The META-SHARE OWL ontology for the interoperability of linguistic datasets on the Web." In European Semantic Web Conference, pp. 271-282. Springer International Publishing, 2015.